Whole Genome Sequencing (WGS) Program
Whole genome sequencing (WGS) is a cutting-edge technology that FDA has put to a novel and public health-promoting use. FDA is laying the foundation for the use of whole genome sequencing to protect consumers from foodborne illness in countries all over the world.
Using Science to
Find the Sources of Outbreaks
(PDF: 544KB)
An Economic Evaluation of the Whole Genome Sequencing Program in the U.S.
Whole genome sequencing reduces illnesses and saves hundreds of millions of dollars each year.
On this page:
- Introduction
- GenomeTrakr: Using Genomics to Identify Food Contamination
- How FDA Uses Whole Genome Sequencing for Regulatory Purposes
- Proactive Applications of Whole Genome Sequencing Technology
Introduction
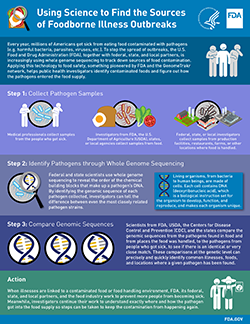
Whole genome sequencing reveals the complete DNA make-up of an organism, enabling us to better understand variations both within and between species. This in turn allows us to differentiate between organisms with a precision that other technologies do not allow. FDA is using this technology to perform basic foodborne pathogen identification during foodborne illness outbreaks and applying it in novel ways that have the potential to help reduce foodborne illnesses and deaths over the long term both in the U.S and abroad.
The most basic application of this technology to food safety is using it to identify pathogens isolated from food or environmental samples. These can then be compared to clinical isolates from patients. If the pathogens found in the food or food production environment match the pathogens from the sick patients, a reliable link between the two can be made, which helps define the scope of a foodborne illness outbreak. This type of testing has traditionally been done using methods such as PFGE, but there are some strains of Salmonella spp. that PFGE is unable to differentiate. Whole genome sequencing performs the same function as PFGE but has the power to differentiate virtually all strains of foodborne pathogens, no matter what the species. Its ability to differentiate between even closely related organisms allows outbreaks to be detected with fewer clinical cases and provides the opportunity to stop outbreaks sooner and avoid additional illnesses.
However the most promising and far reaching public health benefit may come from pairing a foodborne pathogen’s genomic information with its geographic location and applying the principles of evolutionary biology to determine the relatedness of the pathogens. Why? Because, the genomic information of a species of foodborne pathogen found in one geographic area is different than the genomic information of the same species of pathogen found in another area. Knowing the geographic areas that pathogens are typically associated with can be a powerful tool in tracking down the root source of contamination for a food product, especially multi-ingredient food products whose ingredients come from different states or countries. The faster public health officials can identify the source of contamination, the faster the harmful ingredient can be removed from the food supply and the more illnesses and deaths that can be averted.
To realize this goal FDA is spearheading an international effort to build a network of laboratories that can sequence the genomes of foodborne pathogens and then upload the genomic sequence of the pathogen and the geographic location from which the pathogen was gathered into a publicly accessible database. As the size of the database grows, so will its strength as a tool to help focus and speed investigations into the root cause of illnesses.
FDA’s foods program has been utilizing whole genome sequencing since 2008.
GenomeTrakr: Using Genomics to Identify Food Contamination
The FDA Foods Whole Genome Sequencing Staff is coordinating efforts by public health officials to sequence pathogens collected from foodborne outbreaks, contaminated food products, and environmental sources. The genome sequences are archived in an open-access genomic reference database called GenomeTrakr, that can be used: to find the contamination sources of current and future outbreaks; to better understand the environmental conditions associated with the contamination of agricultural products; and to help develop new rapid methods and culture independent tests.
- GenomeTraker Fast Facts
Includes updates on the number of pathogens sequenced - Video overview of how foodborne pathogens are isolated from a food sample and sequenced
- GenomeTrakr: Pushing Back the Frontiers of Outbreak Response
- GenomeTrakr: Transforming Food Safety
- Sharing Whole Genome Sequencing with the World
- FDA Assesses Impact of Whole Genome Sequencing Program
Network estimated to generate hundreds of millions of dollars annually in net health benefits - Whole Genome Sequencing in the New Era of Smarter Food Safety
- More details about GenomeTrakr
How FDA Uses Whole Genome Sequencing for Regulatory Purposes
Genomic data from foodborne pathogens, by itself and in combination with other information, is a robust resource that can help public health officials identify and understand the source of foodborne illness outbreaks. It can be used: to determine which illnesses are part of an outbreak and which are not; to determine which ingredient in a multi-ingredient food is responsible for an outbreak; to identify geographic regions from which a contaminated ingredient may have originated; to differentiate sources of contamination, even within the same outbreak; to link illnesses to a processing facility even before the food product vector has been identified; to link small numbers of illnesses that otherwise might not have been identified as common outbreak; and to identify unlikely routes of contamination.
Read about how FDA has used whole genome sequencing of foodborne pathogens for regulatory purposes.
Proactive Applications of Whole Genome Sequencing Technology
Although public health officials sequence foodborne pathogens after a foodborne illness outbreak or event has occurred, that isn’t the only time they are sequenced and the genomic information sequencing provides can be used for more than just determining the scope of outbreaks and speeding traceback investigations. It can be used: as an industry tool for monitoring ingredient supplies, the effectiveness of preventive and sanitary controls, and to develop new rapid method and culture independent tests; to determine the persistence of pathogens in the environment; to monitor emerging pathogens; and as a possible indicator of antimicrobial resistance.
Read more about proactive applications of whole genome sequencing technology.