BAM Protocol: Simultaneous Confirmation of Listeria species and L. monocytogenes isolates by real-time PCR
Identification of Listeria species and specifically Listeria monocytogenes by real-time PCR allows rapid and accurate confirmation of presumptive isolates. A real-time 5'-nuclease PCR assay has been developed targeting different regions of the iap gene to detect L. monocytogenes as well as Listeria species, including L. monocytogenes, L. innocua, L. ivanovii, L. seeligeri and L. welshimeri. (The identification of Listeria grayi has not been verified with this assay.) This duplex real-time PCR assay is intended for the confirmation of isolates and is performed with the Smart Cycler II (Cepheid, Sunnyvale CA) instrument platform.
- Equipment and Supplies
- SmartCycler II (Cepheid, Sunnyvale, CA) or PCR Thermalcycler capable of performing cycling parameters described below and simultaneous real-time sequence detection of FAM and Texas Red dyes.
- PCR tubes (minimum reaction volume of 25 µl) and trays compatible with PCR thermalcycler.
- Media and Reagents
- Sterile Molecular Grade water
- OmniMix-HS PCR Reagent Beads (Cepheid, Sunnyvale, CA. Also available through Fisher).
- Listeria species and L. monocytogenes iap primers and probes (Table 1).
- Stock and Working (10 µM) solutions can be prepared from commercially synthesized primers with basic desalt purification (Fisher/Genosys or equivalent) by rehydrating with sterile molecular biology grade water to appropriate concentrations.
- Store at -20°C to -70°C non-frost-free freezer
Table 1. Primer/probe sequences for use on SmartCycler II platform (Cepheid, Sunnyvale, CA) based on the sequence of GenBank Accession X52268.
Primers1 Bases 5' → 3' Sequence Lm835F 28 AACTGGTTTCGTTAACGGTAAATACTTA Lm998R 20 TAGGCGCAGGTGTAGTTGCT Lall1055F 24 GTTAAAAGCGGTGACACTATTTGG Lall1163R 31 TTTGACCTACATAAATAGAAGAAGAAGATAA Probes1 Lall1118PFAM 15 6FAM-ATGTCATGGAATAAT-MGB-NFQ Lm918P TxRd 28 TxRd-CTACTACTCAACAAGCTGCACCTGCTGC-IowaBlackRQ 1Primer/Probe name composed of target (Lall = all Listeria species, Lm = L. monocytogenes), 5' base position of oligonucleotide in the respective gene sequence specified in column 2 and forward primer (F), reverse primer (R) or probe (P).
- Control Cultures
- Listeria monocytogenes ATCC 19115
- Listeria innocua ATCC 33090
- Listeria seeligeri ATCC 35967
- Listeria ivanovii ATCC 19119
- Template Preparation
- Prepare from bacterial isolates cultured in Brain Heart Infusion Broth (BHI), 20-24 hr at 35°C or from a colony grown on agar plate suspended in 0.85% saline.
- Transfer 1ml of the suspension to a microcentrifuge tube and centrifuge 12000 × g for 3 min.
- Remove the supernatant and completely resuspend pellet in 1 ml 0.85% NaCl.
- Centrifuge 12000 × g for 3 min.
- Remove the supernatant and resuspend the pellet in 1 ml sterile water.
- Boil templates for 10 min.
- Centrifuge 12,000 × g for 1 minute, remove the supernatant and save as DNA template (This may be frozen, -20°C, for future PCR tests).
- PCR Controls:
- For a positive PCR control include template prepared from L. monocytogenes, such as ATCC #19115 that has both gene targets.
- Always run a no template (water) negative control tube in every run.
- Real-Time PCR assembly and data analysis protocols:
- Reaction assembly
- Prepare a PCR Master Mix from the reaction components and final concentrations listed in Table 2. Keep all thawed reagents and reactions on ice / cooling block.
- Add 24 µl of Master Mix to each SmartCycler tube and cap loosely.
- Add 1 µl of sample or control template and snap cap tightly.
- Briefly centrifuge to bring all liquid to bottom of tube and place in thermalcycler.
- Create run on SmartCycler II. Give each run a unique run name, select Dye set FTTC25, select 2-step PCR protocol as described below and assign appropriate sites on SC block.
Initial Activation 60 sec at 94°C 45 cycles 10 sec at 94°C, (optics off) 45 sec at 60°C, (optics on) - Qualitative data analysis
On SmartCycler II Instrument set the following Analysis Settings for FAM, and TxRd channels. Update analysis settings if they are changed before recording results.
Usage Assay Curve Analysis Primary Threshold Setting Manual Manual Threshold
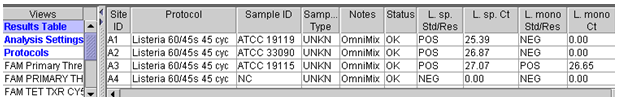
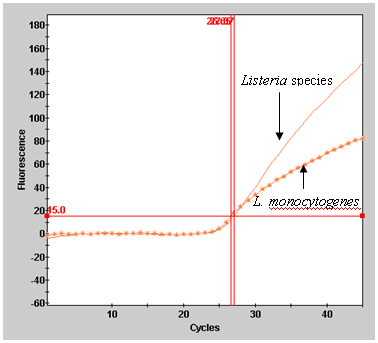
Fluorescence Units15.0 Auto Min Cycle 5 Auto Max Cycle 10 Valid Min.Cycle 3 Valid Max. Cycle 60 Background subtraction ON Boxcar Avg. Cycles 0 Background Min. Cycle 5 Background Max. Cycle 40 Primary fluorescence curves that cross the threshold will be recorded as "POS" and the cycle the sample crossed the threshold will be displayed in the Results Table view. Results can also be viewed graphically. A report can be generated or a screen capture of the results table view can be used to record data (Figure 1).
- Interpretation of Real-Time PCR Results
-
If both gene targets in the FAM and TxRd channels are POS the isolate is L. monocytogenes.
If only the FAM channel is POS the isolate is a Listeria species, not L. monocytogenes.
-
Table 2. Smart Cycler II Protocol Amplification Reaction Components
Volume/rxn Component Final
Concentrationqsi to 25 µl1 Sterile Distilled Water 0.625 µl Primer LallF1055 (10 µM Work Solution) 0.25 µM 0.625 µl Primer LallR1163 (10 µM Work Solution) 0.25 µM 0.625 µl Primer LmF835 (10 µM Work Solution) 0.25 µM 0.625 µl Primer LmR998 (10 µM Work Solution) 0.25 µM 0.25 µl Probe LallP1118FAM(10 µM Work Solution) 0.1 µM 0.25 µl Probe LmP918TxRd (10 µM Work Solution) 0.1 µM 0.5 bead OmniMix-HS 1-5 µl Template (Sample or control) 1Appropriate amount of sterile distilled water is added depending on sample
template volume being used.Figure 1. Example of result output from Smart Cycler II.
A. Results table view for L. ivanovii, ATCC 19119, L. innocua, ATCC 33090 and L. monocytogenes, ATCC 19115.
B. Results graphical view for L. monocytogenes, ATCC 19115.
- Reaction assembly