GenomeTrakr Network
The GenomeTrakr network is the first distributed network of laboratories to utilize whole genome sequencing for pathogen identification. It consists of public health and university laboratories that collect and share genomic and geographic data from foodborne pathogens. The data, which are housed in public databases at the National Center for Biotechnology Information (NCBI), can be accessed by researchers and public health officials for real time comparison and analysis that can speed foodborne illness outbreak investigations and reduce foodborne illnesses and deaths.
In addition to sequencing foodborne pathogens, the GenomeTrakr network may also sequence non-foodborne illness pathogens that have the potential to disrupt the food supply in other ways. An example of such a pathogen is the SARS-CoV-2 (COVID-19) virus, which has demonstrated its ability to disrupt the nation’s food supply infrastructure by causing widespread illness, including among food production and foodservice workers.
- Fast Facts - Includes updates on the number of pathogens sequenced
- GenomeTrakr: Revealed
- GenomeTrakr: Pushing Back the Frontiers of Outbreak Response
- GenomeTrakr: Transforming Food Safety
- Access GenomeTrakr Data via NCBI’s Pathogen Detection Portal
- Sharing Whole Genome Sequencing with the World
- Whole Genome Sequencing in the New Era of Smarter Food Safety
- WGS and GenomeTrakr Q&A (PDF: 170KB)
- Joining and Using the GenomeTrakr Network
Real Time Food Safety Applications of the GenomeTrakr Network
Collaboration with NCBI
With its mission of providing publicly accessible information and tools that enhance understanding of how genetic processes impact health and disease, NCBI has played a key role in the development and success of GenomeTrakr. NCBI collects, houses, and analyzes foodborne pathogen whole genome sequences to reveal their relatedness to one and other. The genomic sequences, supporting metadata, and the results of NCBI’s analyses are presented in easy-to-use web interfaces that can be used by public health officials, researchers, and members of industry to inform their decisions. Since 2013 the genomic information NCBI houses has been used by public health officials to support more than 1,643 actions intended to protect consumers from foodborne illness. Examples of public health actions supported by genomic data analyzed by NCBI include investigations of multistate outbreaks of: Listeria monocytogenes infections linked to Frozen Supplemental Shakes (2025); Salmonella Enteritidis infections linked to eggs (2025); and Salmonella Anatum infections linked to mung and moth bean sprouts (2025). WGS findings have also prompted FDA to issue numerous warning letters and companies to recall products, including pet food. GenomeTrakr data can be accessed for review and analysis via NCBI’s Pathogen Detection portal.
Foodborne Pathogen Sequencing
GenomeTrakr labs perform whole genome sequencing on Salmonella, Listeria, E. coli, Campylobacter, Vibrio, Cronobacter, etc. isolates, as well as parasites and viruses, to leverage public health benefits that can be derived from the open sharing of their genomic information. In the U.S., food and environmental samples are sequenced by FDA, USDA, state, academic, and other GenomeTrakr partners, while CDC and its affiliated state PulseNet laboratories are the primary network contributors of human clinical isolate sequences. Public health, university, and other labs outside the U.S. contribute sequences from food, environmental, and clinical samples. The genomic sequences and corresponding collection information for the samples are publicly available via NCBI’s Pathogen Detection portal.
Wastewater Surveillance for SARS-CoV-2 Variants
Studies have shown that SARS-CoV-2 (COVID-19) variants of concern from wastewater can be identified 1-2 weeks prior to being detected in clinical samples from the same area, making wastewater surveillance useful for detecting and monitoring SARS-CoV-2 in the population. Accordingly, FDA has developed the capacity to sequence SARS-CoV-2 RNA from wastewater samples. By monitoring for both known and unknown variants of the virus, GenomeTrakr labs are providing information about the evolution of the virus, which is critical to evaluating the effectiveness of FDA-regulated COVID-19 vaccines, therapeutics, and diagnostics. Geographic regions with high percentages of agricultural and food facility workers, whose health is critical for stability of the U.S. food supply chain, are of interest. Resulting sequence data are being submitted in real time to public repositories where active monitoring for circulating variants of concern takes place. Additional information about the project can be found at Wastewater Surveillance for SARS-CoV-2 Variants.
Sequencing of Historical Samples
Many public health laboratories have pathogen isolates from past outbreaks stored in their freezers. These isolates hold a treasure trove of genomic information waiting to be unlocked by whole genome sequencing. FDA encourages those labs to sequence those isolates and upload the genomic information to the GenomeTrakr database at NCBI.
Joining and Using the GenomeTrakr Network
For information about joining the GenomeTrakr network as a sequencing lab, providing isolates to a current member lab for sequencing, or using the GenomeTrakr database as a research tool, please contact FDA at GenomeTrakr@fda.hhs.gov.
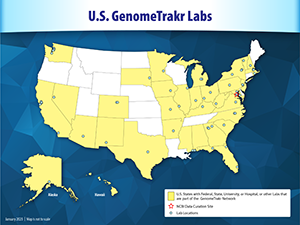
Contributing Labs:
U.S. FDA Labs
Gulf Coast Seafood Laboratory, Dauphin Island, AL
Arkansas Regional Laboratory, Jefferson, AR
San Francisco District Laboratory, Alameda, CA
Pacific Regional Laboratory—Southwest, Irvine, CA
Denver District Laboratory, Denver, CO
Southeast Regional Laboratory, Atlanta, GA
CFSAN Research Laboratories at Moffett Campus, Bedford Park, IL
CFSAN Molecular Methods and Subtyping Lab, College Park, MD
ORA-CFSAN Method Development and Validation Laboratory at MOD1, Laurel, MD
Winchester Engineering & Analytical Center, Winchester, MA
Northeast Regional Laboratory, Jamaica, NY
Forensic Chemistry Center, Cincinnati, OH
Pacific Regional Laboratory—Northwest, Bothell, WA
U.S. State and University Labs
Alaska State Public Health Laboratory, Anchorage, AK
Arizona Department of Health Services with the Translational Genomics Research Institute, Phoenix, AZ*
California Department of Public Health, Richmond, CA*
Colorado Department of Public Health and Environment, Denver, CO*
NOVA Southeastern University, Fort Lauderdale, FL
Florida Department of Health, Jacksonville, FL
Florida Department of Agriculture and Consumer Services, Tallahassee, FL*
Hawaii Department of Health, Pearl City, HI
Indiana State Department of Health, Indianapolis, IN*
State Hygienic Laboratory at the University of Iowa, Coralville, IA
Kentucky Cabinet for Health and Family Services, Frankfort, KY*
Maryland Department of Health and Mental Hygiene, Baltimore, MD*
Joint Institute for Food Safety and Applied Nutrition, College Park, MD**
Massachusetts State Public Health Laboratory, Jamaica Plain, MA*
Michigan Department of Agriculture and Rural Development, East Lansing, MI*
Michigan Department of Health and Human Services, Lansing, MI*
Minnesota Department of Health, Saint Paul, MN*
Missouri Department of Health and Senior Services, Jefferson City, MO*
Nevada Department of Health and Human Services with University of Nevada - Reno, Reno NV*
New Hampshire Department of Health and Human Services, Concord, NH*
New Jersey State Department of Agriculture, Animal Health Diagnostic Laboratory, Ewing, NJ*
New Jersey Department of Health, Trenton, NJ*
New Mexico State University, Food Safety Laboratory, Las Cruces, NM*
New York State Department of Agriculture & Markets, Albany, NY*
New York State Department of Health - Wadsworth Center, Albany, NY*
Cornell University, Ithaca, NY
North Carolina Department of Agriculture and Consumer Services, Raleigh, NC*
North Carolina State University, College of Veterinary Medicine, Raleigh, NC*
Animal Diseases Diagnostic Laboratory, Ohio Department of Agriculture, Reynoldsburg, OH*
Pennsylvania Department of Health with Pennsylvania State University, University Park, PA*
Rhode Island Department of Health, Providence, RI*
South Carolina Department of Health and Environmental Control, Columbia, SC*
South Dakota Department of Health with South Dakota State University, Brookings, SD*
Texas Department of State Health Services, Austin, TX*
Vermont Agency of Human Services, Colchester, VT*
Virginia Department of Health, Richmond, VA*
University of Washington, Seattle, WA
Washington State Department of Agriculture, Olympia, WA*
Washington State Department of Health Public Health Laboratories, Shoreline, WA*
West Virginia Department of Agriculture, Charleston, WV*
U.S. Hospital Labs
Brigham and Women's Hospital, Boston, MA
U.S. Department of Agriculture (USDA) Food Safety Inspection Service (FSIS) Field Services Labs
Western Laboratory, Albany, CA
Eastern Laboratory, Athens, GA
Midwestern Laboratory, St. Louis, MO
Centers for Disease Control and Prevention (CDC) Labs
Enteric Diseases Laboratory, Atlanta, GA
National Oceanic and Atmospheric Administration (NOAA) Labs
Cooperative Oxford Lab, Oxford, MD
National Seafood Inspection Laboratory, Pascagoula, MS
Other Labs located in the U.S.
Gladstone Institutes, San Francisco, CA
IEH Laboratories & Consulting Group, Lake Forest Park, WA
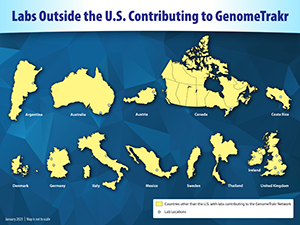
Labs located outside of the U.S.
INEI-ANLIS “Carlos Malbran Institute,” Buenos Aires, Argentina
Doherty Institute, Melbourne, Australia
Institute of Clinical Pathology and Medical Research, Westmead Hospital, Sydney, Australia
University of New South Wales, Sydney, Australia
Austrian Agency for Health and Food Safety, Vienna, Austria
Health Canada, Ottawa, Canada
Laboratoire de sante publique du Quebec, Sainte-Anne-de-Bellevue, Canada
Instituto Costarricense de Investigación y Enseñanza en Nutrición y Salud (INCIENSA), Tres Rios, Cartago, Costa Rica
Danish Technical University, Kongens Lyngby, Denmark
Statens Serum Institute, Copenhagen, Denmark
Justus Liebig University, Giessen, Germany
University Hospital Muenster, Muenster, Germany
Alagappa University, Karaikudi, India
Cochin University of Science and Technology, Kochi, India
Dr. M.G.R Fisheries College and Research Institute, Thalainayeru, India
Centre for Food Safety, University College Dublin, Dublin, Ireland
National Salmonella, Shigella, and Listeria Reference Laboratory, Galway, Ireland
Istituto Zooprofilattico Sperimentale dell'Abruzzo e del Molise "Giuseppe Caporale" (IZSAM), Teramo, Italy
Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria (SENASICA), Mexico City, Mexico
Livsmedelsverket (Swedish Food Agency), Uppsala, Sweden
Ministry of Public Health, Mueang Nonthaburi, Thailand
University of Birmingham, Birmingham, United Kingdom
NHS Lothian, Edinburgh, United Kingdom
Public Health England, London, United Kingdom
University of Oxford, Oxford, United Kingdom
Animal and Plant Health Agency, Surrey, United Kingdom
Food Environmental Research Agency (Fera), York, United Kingdom
Collaborations with Independent Academic Researchers:
In addition to state labs, university labs, federal labs, and labs located outside of the U.S., the GenomeTrakr network has collaborative relationships with a number of independent researchers, including graduate students, who supply foodborne pathogen isolates and/or perform whole genome sequencing on the isolates.
Data Curation:
Data curation and bioinformatic analyses and support are provided by the National Center for Biotechnology Information (NCBI) in Bethesda, MD.
* Receives funding from FDA through FDA’s Laboratory Flexible Funding Model (LFFM) Cooperative Agreement Program
** Receives funding from FDA through a mechanism other than the LFFM Cooperative Agreement Program